Rebuilding the bacterial cell division machinery in vitro
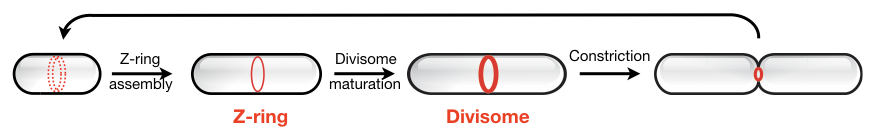
Even though we probably have a complete list of proteins involved in bacterial cell division, how these molecular players are able to self-organize into a functional machinery at the right place and time is not clear.
We are aiming to understand the mechanism of bacterial cytokinesis by rebuilding the cell division machinery from purified components in vitro.
Using this approach, we have been able to demonstrate the ability of FtsZ to form treadmilling filaments in vitro:
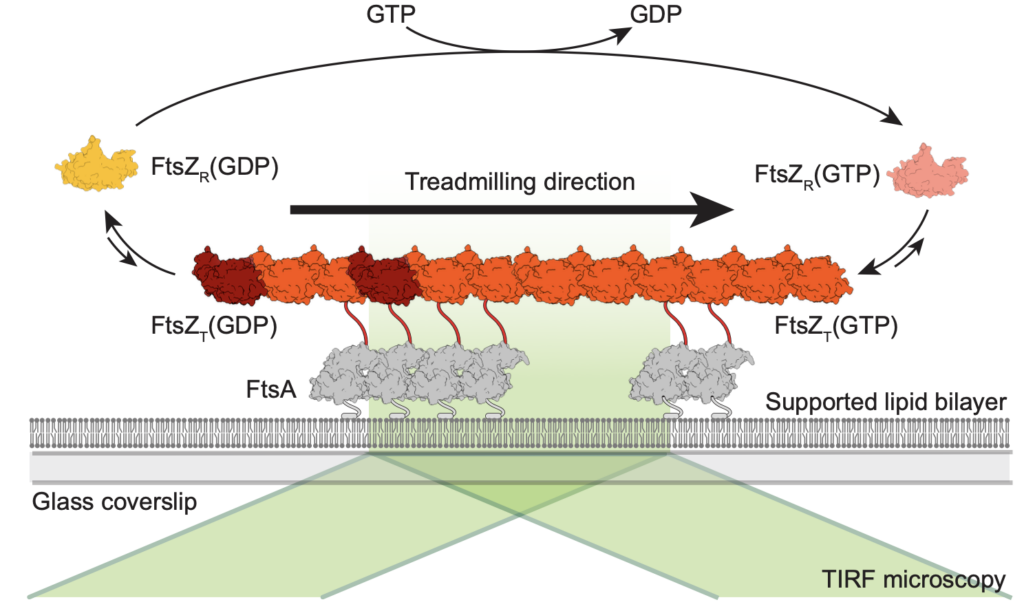
Now, we are using this assay to answer the following questions:
How do FtsZ associated proteins modulate the organization of FtsZ filaments?
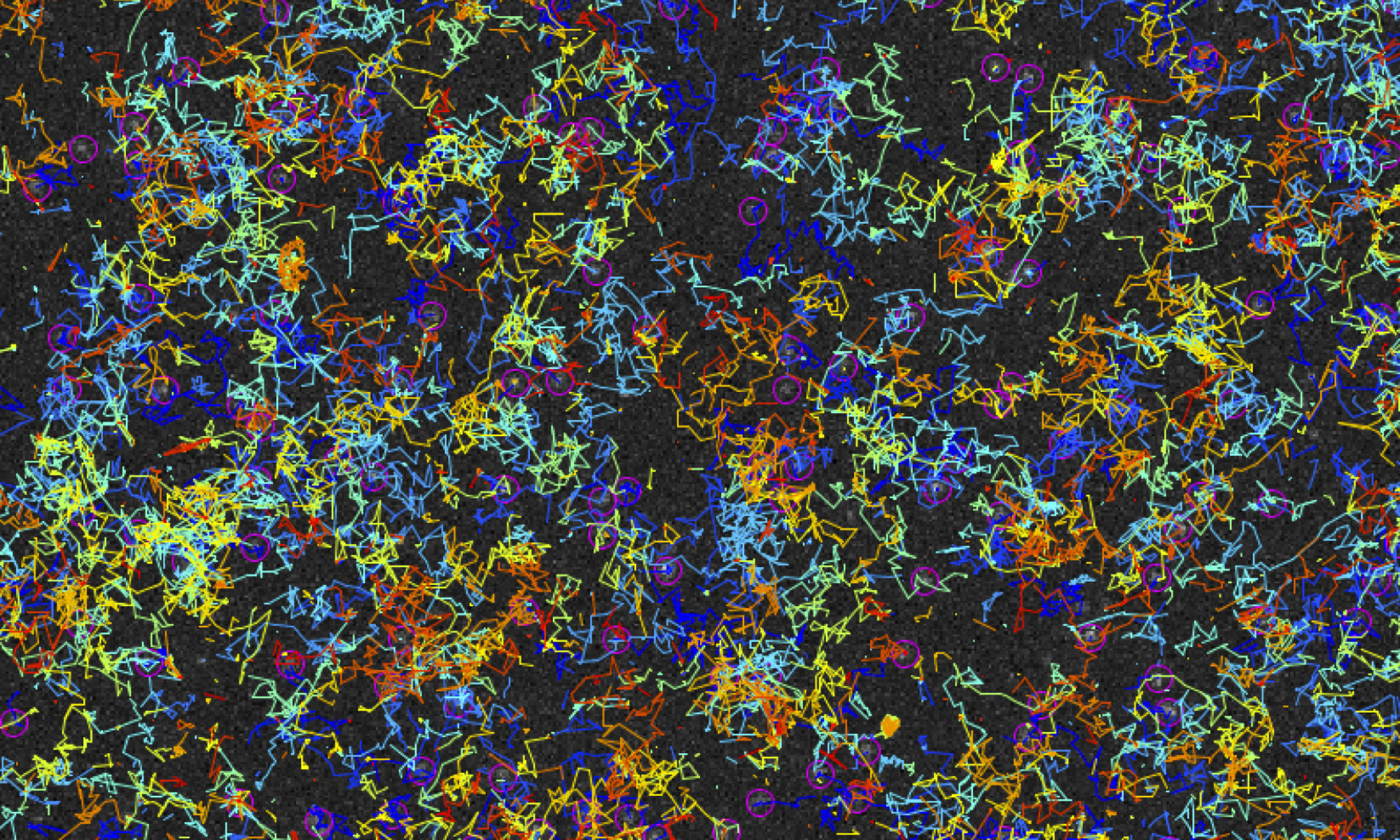
The Z-ring has long been known to act as a dynamic scaffold for the assembly of the bacterial cell division machinery. What governs the architecture, dynamics and stability of this cytoskeletal structure was unclear, but FtsZ-associated proteins (Zaps) had been suggested to play an important role. We used in vitro reconstitution to study the influence of the well-conserved protein ZapA on the self-organization of FtsZ and FtsA on lipid membranes. Combining high-resolution fluorescence microscopy with novel quantitative image analysis (Caldas et al., 2020), we found that ZapA aligns FtsZ filaments and stabilizes filament bundles in a highly cooperative manner. Despite its strong influence on their large-scale organization, we found that ZapA binds only transiently to FtsZ filaments and has no effect on their treadmilling velocity. Our results led to a new model for how FtsZ-associated proteins can increase the precision and stability of bacterial cell division. Furthermore, we propose that the ultrasensitive response of FtsZ filaments to ZapA is important for the directional maturation of the cell division machinery (Caldas et al., 2019).
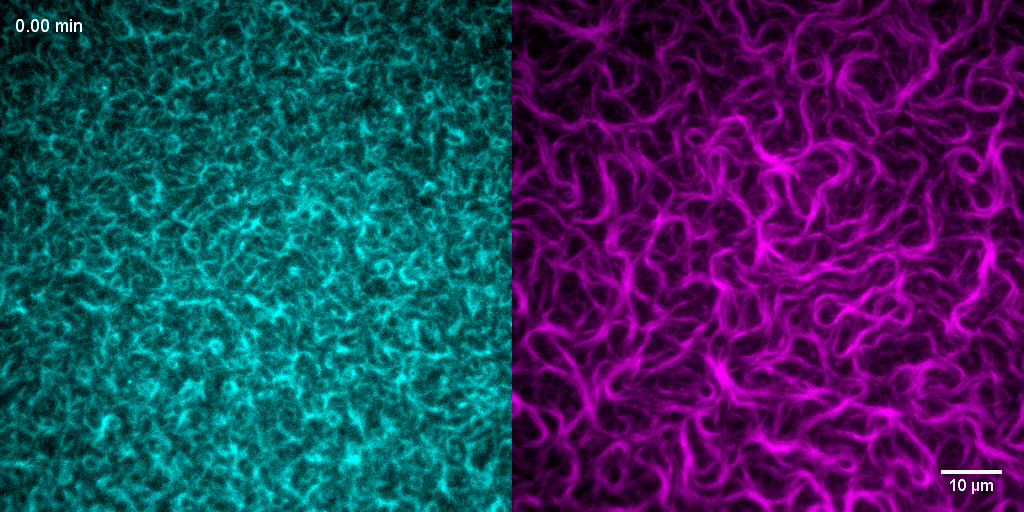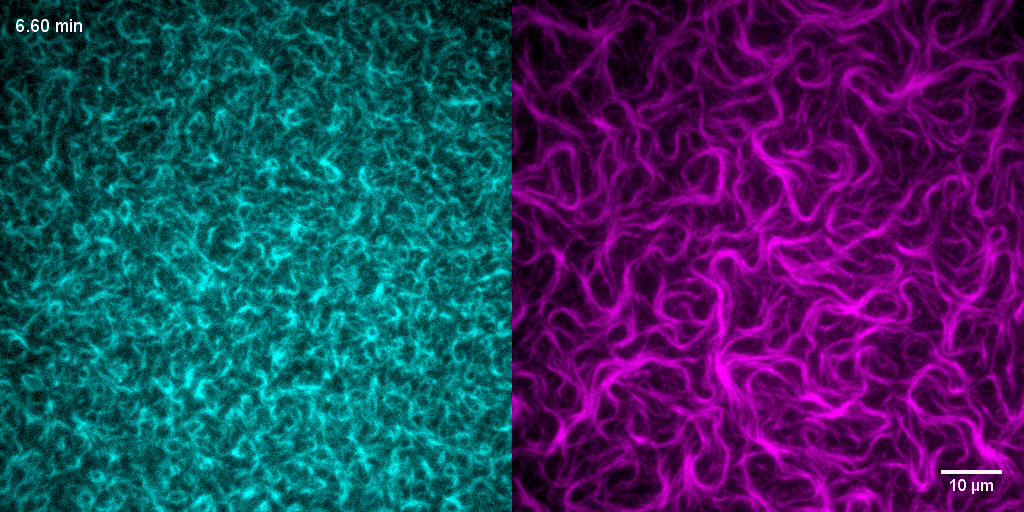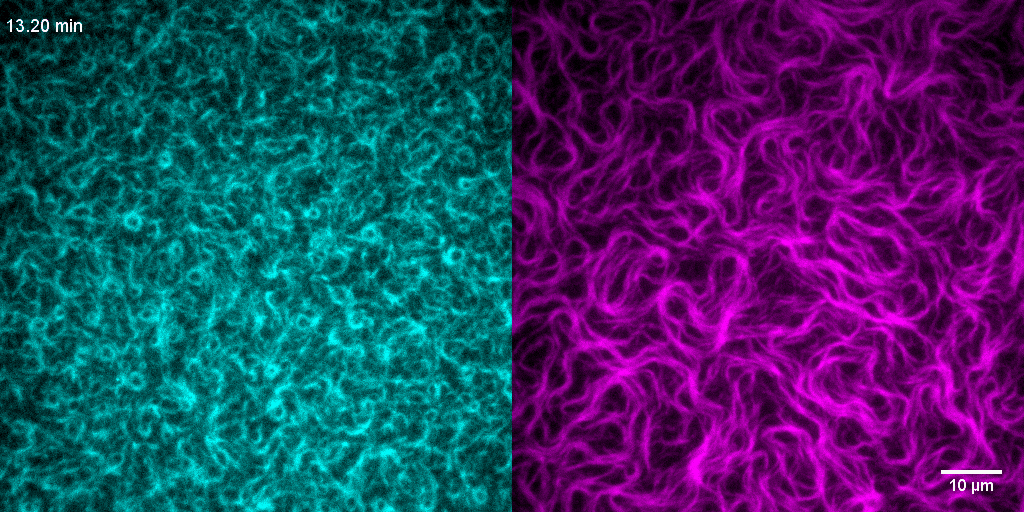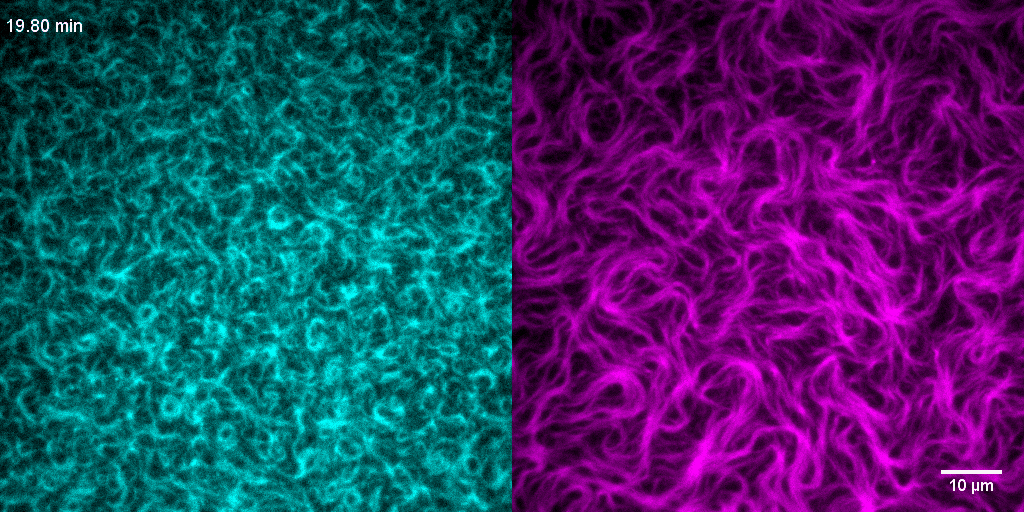
How do treadmilling FtsZ filaments organize cell division proteins in space and time?
The Z-ring not only defines the site of cell division, treadmilling of FtsZ filaments in this structure was also found to actively distribute cell wall synthesis around the cell thereby promoting the integrity of the new formed cell poles (Yang et al., 2017; Bisson-Filho et al., 2017). To understand how FtsZ filament dynamics are coupled to cell wall synthesis, we reconstituted part of the bacterial cell division machinery using its purified components FtsZ, FtsA and truncated transmembrane proteins essential for cell division (Baranova et al., 2020). We found that the membrane-bound cytosolic peptides of FtsN and FtsQ co-migrated with treadmilling FtsZ-FtsA filaments. Remarkably, despite their directed motion on the ensemble level, individual peptides showed random diffusion and transient confinement. Our findings reveal that divisome proteins dynamically follow treadmilling FtsZ filaments by a diffusion-and-capture mechanism, which can help to establish a moving signaling zone at the division site in vivo. This work represents an important experimental demonstration for how transient interactions can lead to the self-organization of complex biological structures.
Current projects
Currently, we are using our in vitro reconstitution approach to answer the following questions:
- How do treadmilling FtsZ filaments distribute cell division proteins?
- How do FtsZ filaments laterally interact to form a coherent structure?
- How does FtsA interact with other cell division proteins?
People working on this project
Philipp, Roman, Marko, Xing, Olesia
Collaborators (past and present)
- Waldemar Vollmer (Newcastle University, UK)
- Jan Brugués (TU Dresden, Germany)
- Anđela Šarić (ISTA, Klosterneuburg, Austria)
- Edouard Hannezo (ISTA, Klosterneuburg, Austria)
- Sonja-Verena Albers (University of Freiburg, Germany)
- Richard Wong (WPI Nano Life Science Institute, Kanazawa University, Japan)
Relevant publications
Zuzana Dunajova, Batirtze Prats Mateu, Philipp Radler, Keesiang Lim, Dörte Brandis, Philipp Velicky, Johann Georg Danzl, Richard W Wong, Jens Elgeti, Edouard Hannezo, Martin Loose
Chiral and nematic phases of flexible active filaments
Nature Physics (2023).
P. Radler, N. Baranova, P. Caldas, C. Sommer, M. López-Pelegrín, D. Michalik, M. Loose.
In vitro reconstitution of Escherichia coli divisome activation
Nature Communications (2022), vol. 13, 2635
N. Baranova, P. Radler, V. M. Hernandez-Rocamora, C. Alfonso, M. Lopez-Pelegrin, G. Rivas, W. Vollmer, M. Loose.
Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins.
Nature Microbiology (2020), vol. 5(3):407-417.
P. Caldas, M. Lopez-Pelegrin, D.J.G. Pearce, N.B. Budanur, J. Brugues and M. Loose.
Cooperative Ordering of Treadmilling Filaments in Cytoskeletal Networks of FtsZ and Its Crosslinker ZapA.
Nature Communications (2019), vol. 10 (1), 5744.
P. Caldas, P. Radler, C. Sommer and M. Loose.
Computational analysis of filament polymerization dynamics in cytoskeletal networks.
Methods in Cell Biology (2020), vol. 158:145-161.
Funding

This research project was funded by the European Research Council (ERC) with a ERC Starting Grant (“Self-organization of the bacterial cell”, Grant No. 679239).

This research project has received funding from the Austrian Science Fund (FWF) StandAlone P34607.